Tags
CDK, Isomorphism, Java, Similarity, SMSD, substructure, VF2
Updated (17 March 2010): Gilleain and myself were able to code an ultrafast VF2 algorithm for Isomorphism substructure search (permutation bug is resolved). The code is ported or re-factored from Chemkit and I have further optimised the code at our end. The previous VF2 code in the SMSD is more MCS oriented which was a big compromise in terms of speed while performing a substructure search. I have mentioned these issues in a previous blog. The whole idea of a substructure search is to answer a boolean question i.e. is A a subgraph/substructure of B ? If the answer is true then matching atoms are reported. On the other hand MCS where it’s about reporting the commonality between A and B.
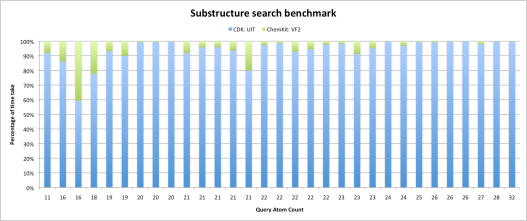
Below is the benchmark graph which speaks about the performance (30 queries were subjected to 15030 targets, same as my previous benchmark test cases) of the VF2.
- X-axis: 30 test cases subjected to 15030 targets (results are sorted in ascending order of the query atom count). Y- axis: total percentage of time spent in calculating the substructures.
As is apparent from the benchmark graph, VF2 takes less than 10-20% of the total time (in most cases) for performing substructure searches (results are sorted in ascending order of the query atom count). So as for now we have an implementation which at last outperforms UIT (CDK code) in terms of speed for automorphic graphs. The performance of the VF2 improves as the query atom count (graph size) increases. We might need a few changes in the CDK AtomContainer class itself to further improve the speed/performance but this will involve further discussions with CDK developers.

*very* impressive. Nice work! Is there anything weird about structure 12, that makes it an outlier?
BTW, I think you swapped the description of the X & Y axes
Thanks! I have updated the fig. as there was an indexing bug.
Now the figure legends are also fixed 🙂
So, what’s this 12th substructure like?
Sorry that was an indexing bug, sorted out!
Actually, can you also create a plot showing how they scale with respect to substructure size, with the lines in a plot of absolute time versus substructure size? Or perhaps some complexity measure, if you think that is more important…
Thanks! I have updated the figure and the results are sorted out in ascending order of the query atom count. VF2 is more consistent with larger graphs.
Hi Asad, very nice. Can you please post a link to your test molecules with mol files. I guess you worked with open data 🙂 Cheers, Chris
Hi Chris, I have used the data set provided by Rajarshi from his benchmark test conducted on the CDK fingerprints. These dataset contains AID’s 466 and 548 (which were cleaned locally by Rajarshi). I will gist them if Rajarshi agrees 🙂
The advantage of a blog is that you can ‘publish’ results immediately.
The DISadvantage of a blog is that you can ‘publish’ results immediately.
The VFLib code is not working, except for the trivial case of the identity isomorphism. In other words, it seems to be checking if two molecules have identical bond orderings. Naturally enough, this is extremely fast.
I’m not saying that the code is not fast – it certainly should be – but getting the answer right is as important as getting it quickly 🙂
Yes, you are right.. test results with permutations are falling off. I guess we need to check the backtracking part. Shouldn’t be a rocket science as the VF2 lib version in the SMSD works perfectly 🙂
Please test the latest code from the github (Isomorphism and permutations are working)… all fixed!
AND the updated graph on the above blog is as good as the old!
…its still cruising :-).